Biomedical text is dense with critical information—gene names, chemical compounds, diseases, species—but extracting that information manually is time-consuming and error-prone. Enter HunFlair, a specialized Named Entity Recognition (NER) tool built into the widely used Flair NLP framework. Designed specifically for the biomedical domain, HunFlair delivers state-of-the-art accuracy, broad entity coverage, and remarkable ease of use, enabling researchers, clinicians, and developers to extract structured knowledge from unstructured biomedical text—without deep NLP expertise or weeks of model tuning.
HunFlair isn’t just another NER model; it’s a pre-trained, plug-and-play solution that consistently outperforms other leading biomedical NER tools by an average of 7.26 percentage points. Whether you’re parsing scientific publications, clinical trial reports, or electronic health records, HunFlair offers a reliable way to identify key biomedical entities with minimal setup.
Why HunFlair Was Built
General-purpose NER systems often struggle with the unique vocabulary, abbreviations, and syntax of biomedical literature. Existing biomedical NER tools, while accurate, frequently require complex installation, custom dependencies, or extensive configuration—creating barriers for non-experts.
HunFlair was created to solve this: an easy-to-use, highly accurate, and robust NER tagger that works out of the box for biomedical texts across genres and writing styles. Built on Flair—a PyTorch-based NLP framework known for its clean API and powerful embeddings—HunFlair inherits modularity and scalability while specializing in biomedical entity recognition.
Key Strengths That Set HunFlair Apart
Unmatched Accuracy
HunFlair achieves state-of-the-art performance on multiple benchmark datasets for biomedical NER. Its architecture leverages contextual string embeddings and transformer-based representations fine-tuned on diverse biomedical corpora, allowing it to generalize well across documents from journals, patents, and clinical notes.
Broad Biomedical Entity Coverage
Unlike narrow NER tools that recognize only genes or only diseases, HunFlair supports multiple entity types simultaneously, including:
- Genes and proteins
- Chemicals and drugs
- Diseases and disorders
- Species
This multi-label capability reduces the need to run multiple models or stitch together disparate tools.
Simplicity Meets Power
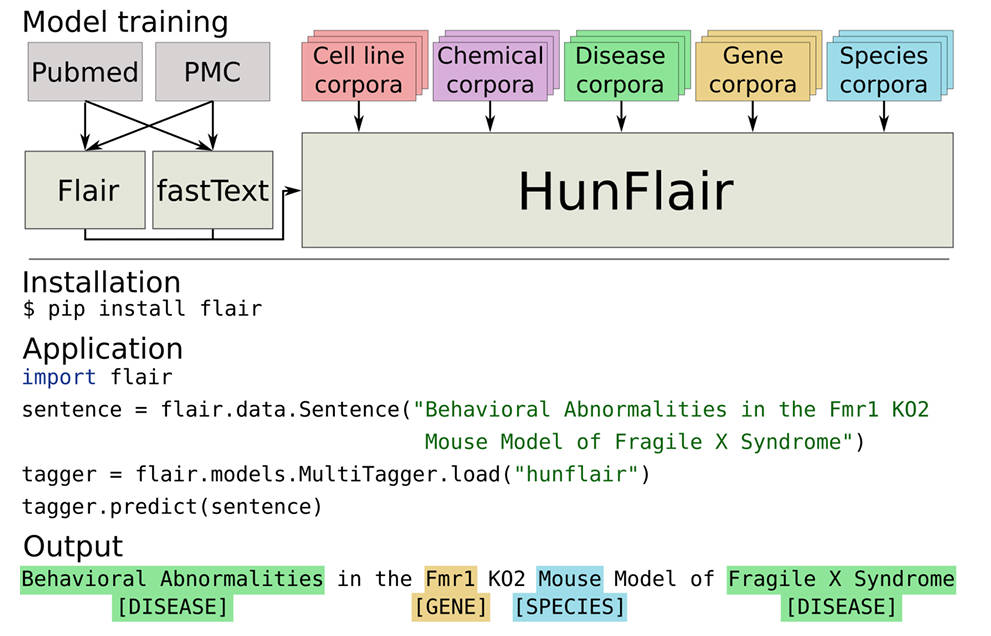
You don’t need to train models, manage GPU resources, or preprocess text manually. HunFlair is pre-trained, ready to run, and integrates seamlessly into Python workflows. Installation takes one command, and inference takes four lines of code.
Perfect Use Cases for HunFlair
HunFlair excels in scenarios where structured extraction from unstructured biomedical text is essential:
- Literature mining: Automatically extract entities from PubMed abstracts or full-text research papers to build knowledge graphs or support systematic reviews.
- Clinical data analysis: Identify diseases, medications, and procedures in de-identified clinical notes or discharge summaries.
- Drug discovery pipelines: Map chemical compounds and gene interactions from scientific databases or patents.
- Biocuration support: Assist human annotators by pre-labeling documents, significantly speeding up manual curation.
If your work involves turning biomedical text into actionable data, HunFlair removes the NLP bottleneck.
Getting Started Is Effortless
HunFlair’s biggest advantage may be its accessibility. Here’s how simple it is to run biomedical NER:
from flair.data import Sentence
from flair.nn import Classifier
sentence = Sentence("BRCA1 mutations increase the risk of breast cancer.")
tagger = Classifier.load('hunflair') # loads the pre-trained HunFlair model
tagger.predict(sentence)
print(sentence)
Output:
Sentence: "BRCA1 mutations increase the risk of breast cancer." - ["BRCA1"/GENE, "breast cancer"/DISEASE]
That’s it. No Docker containers, no environment conflicts, no hyperparameter tuning. Just install with pip install flair (Python 3.9+ required) and start extracting entities.
Behind the scenes, HunFlair uses a sophisticated ensemble of embeddings—including Flair embeddings and biomedical BERT variants—but you never need to manage that complexity.
Limitations and Practical Considerations
While HunFlair is powerful, it’s important to understand its boundaries:
- Domain-specific: It is optimized for biomedical text. Performance on general news, social media, or legal documents will likely be suboptimal.
- Language support: The current HunFlair model is trained primarily on English biomedical text. Non-English use cases may require additional adaptation.
- Text style sensitivity: While robust across scientific writing styles, extremely noisy or informal clinical shorthand (e.g., unstructured physician notes with heavy abbreviations) may reduce accuracy.
- Python requirement: Requires Python 3.9 or newer, which could be a constraint in legacy environments.
These aren’t flaws—they reflect HunFlair’s deliberate focus on delivering best-in-class performance for a well-defined problem.
Is HunFlair Right for Your Project?
Choose HunFlair if:
- You need high-accuracy biomedical NER without investing in model development.
- Your team includes biologists, clinicians, or data scientists—not NLP specialists.
- You want a maintainable, lightweight solution that integrates into existing Python pipelines.
Avoid HunFlair if:
- Your text is non-biomedical (e.g., news, finance, legal).
- You require fine-grained control over model architecture without retraining capabilities.
- You’re working exclusively with non-English biomedical texts not covered by the model.
For the right use case, HunFlair isn’t just convenient—it’s transformative. It turns a historically complex NLP task into a simple function call, accelerating research and development across the life sciences.
Summary
HunFlair redefines what’s possible in biomedical information extraction. By combining state-of-the-art accuracy, multi-entity support, and extreme ease of use, it empowers non-NLP experts to unlock insights from complex scientific text. With a single installation command and four lines of code, you gain access to one of the most accurate biomedical NER systems available—free, open-source, and ready to deploy. If your work touches biomedical literature, HunFlair deserves a place in your toolkit.