Medical image segmentation—the process of delineating organs, tumors, or tissues in scans like MRI or dermoscopic images—is a foundational task in clinical diagnostics and research. Traditional deep learning approaches, such as U-Net variants, have dominated this space for years. However, recent advances in generative modeling have opened new doors. Enter MedSegDiff, a diffusion-based framework specifically engineered for high-precision medical segmentation. Its latest iteration, MedSegDiff-V2, pushes performance further by thoughtfully integrating Vision Transformers (ViTs) into the diffusion pipeline—not as a simple add-on, but through a novel architectural design that actually improves results.
What makes MedSegDiff compelling isn’t just its technical novelty, but its practicality: it delivers state-of-the-art accuracy across more than 20 diverse medical segmentation tasks while remaining adaptable to new datasets with minimal code changes. For teams evaluating tools for medical AI—whether in academia, startups, or healthcare institutions—MedSegDiff offers a powerful yet accessible alternative to conventional segmentation models.
Why Diffusion Models for Medical Segmentation?
Diffusion Probabilistic Models (DPMs) gained fame through image generation (e.g., Stable Diffusion), but their ability to model complex data distributions also makes them well-suited for structured prediction tasks like segmentation. Unlike deterministic models that output a single segmentation mask, diffusion models can capture uncertainty and generate high-quality predictions by iteratively refining noisy inputs.
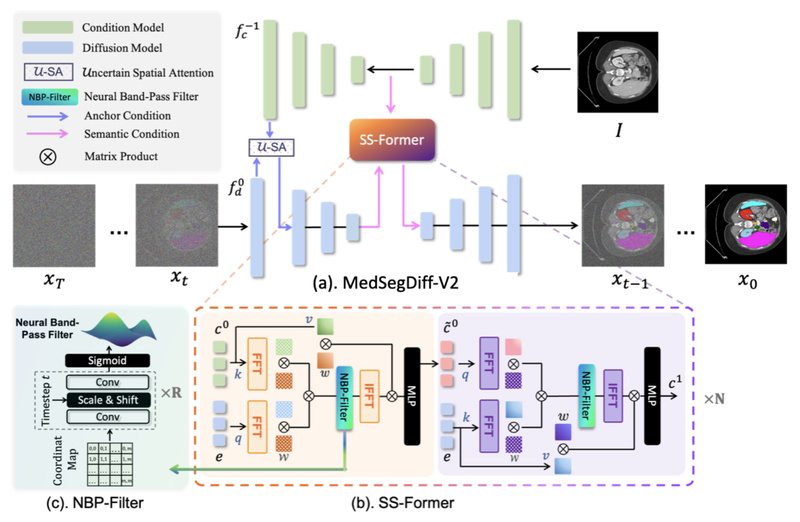
Early attempts to apply DPMs to medical segmentation relied on U-Net backbones. While effective, they left room for improvement—especially in modeling long-range spatial dependencies common in 3D scans or irregular lesion boundaries. MedSegDiff-V2 addresses this by rethinking how transformers interact with the diffusion process. Rather than naively replacing U-Net blocks with ViT layers (which the authors found degraded performance), they designed a coherent Transformer-based diffusion framework that preserves the strengths of both paradigms.
Key Strengths of MedSegDiff-V2
Proven Performance Across Modalities
MedSegDiff-V2 has been rigorously evaluated on 20+ medical image segmentation benchmarks spanning various modalities—MRI (BraTS2020), dermatoscopic images (ISIC), retinal scans, and more. It consistently outperforms prior state-of-the-art methods, a fact validated by its acceptance at top-tier conferences: MIDL 2023 for the original MedSegDiff and AAAI 2024 for MedSegDiff-V2 (which was later named a Most Influential Paper at AAAI-24).
Fast Inference with DPM-Solver
A common criticism of diffusion models is their slow sampling—often requiring hundreds or thousands of steps. MedSegDiff integrates DPM-Solver, enabling high-quality segmentation in as few as 20 steps (down from 1000) without significant quality loss. This makes clinical deployment far more feasible, especially in time-sensitive scenarios.
Flexible and Extensible Architecture
The codebase is modular and well-documented. Swapping datasets doesn’t require overhauling the model—just implementing a new data loader (examples for ISIC and BraTS are included). This lowers the barrier for research teams working with proprietary or niche imaging data.
Tunable Configurations for Diverse Hardware
Whether you’re running experiments on a single 24GB GPU or a multi-GPU cluster, MedSegDiff scales gracefully:
- Lightweight mode:
--num_channels 128,--batch_size 8 - High-performance mode (MedSegDiff++):
--num_channels 512,--num_res_blocks 12,--batch_size 64
This flexibility ensures the framework can be adapted to both resource-constrained labs and well-funded institutions.
Real-World Use Cases
Brain Tumor Segmentation (BraTS2020)
MedSegDiff handles multi-sequence MRI inputs (T1, T2, FLAIR, T1ce) to segment tumor subregions with high fidelity. The framework processes 3D volumes slice-by-slice but maintains consistency through careful preprocessing and postprocessing pipelines included in the repository.
Melanoma Detection (ISIC Dataset)
For 2D dermoscopic images, MedSegDiff accurately outlines skin lesions—critical for early melanoma diagnosis. The official example provides end-to-end scripts for training, sampling, and evaluation using standard metrics.
Beyond Standard Benchmarks
Because the architecture is modality-agnostic, MedSegDiff has been successfully applied to retinal vessel segmentation, lung nodule delineation, and prostate MRI analysis. The project actively encourages community contributions of new dataloaders to expand its reach.
Getting Started: Training, Sampling, and Evaluation
The repository includes clear, ready-to-run examples. Here’s how to replicate results on the ISIC dataset:
- Organize your data in the prescribed folder structure (train/test images and masks).
- Train with:
python scripts/segmentation_train.py --data_name ISIC --data_dir ./data/ISIC --out_dir ./outputs --image_size 256 --num_channels 128 --lr 1e-4 --batch_size 8
- Sample predictions using an ensemble of 5 stochastic runs for robustness:
python scripts/segmentation_sample.py --data_name ISIC --model_path ./outputs/model.pt --num_ensemble 5
- Evaluate against ground truth:
python scripts/segmentation_env.py --inp_pth ./results --out_pth ./data/ISIC/Test/ISBI2016_ISIC_Part1_Test_GroundTruth
For custom datasets, follow the pattern in isicloader.py or bratsloader.py. The maintainers welcome pull requests—this collaborative ethos helps the framework evolve with real-world needs.
Practical Considerations and Limitations
While powerful, MedSegDiff isn’t a “set-and-forget” solution:
- Training dynamics are non-standard: Loss may plateau early, yet segmentation quality continues to improve. Rely on validation metrics—not just training loss—to gauge convergence.
- Hyperparameter sensitivity: Performance depends on careful tuning of architecture and diffusion parameters. The documentation provides sensible defaults, but optimal settings may vary by task.
- GPU memory demands: The high-capacity MedSegDiff++ configuration requires substantial VRAM (~64 GB with batch size 64). Most users will start with the lightweight variant.
- Not fully automated: You’ll need to handle data preprocessing (e.g., NIfTI to slice conversion for BraTS) and postprocessing (e.g., 3D mask reconstruction). Scripts are provided, but domain knowledge helps.
Importantly, the team behind MedSegDiff emphasizes transparency: known issues are documented in the TODO list, and bug fixes are regularly released based on community feedback.
Summary
MedSeg_diff and its evolution, MedSegDiff-V2, represent a significant leap in diffusion-based medical image segmentation. By unifying the generative power of DPMs with the global context awareness of transformers—through a purpose-built architecture rather than a superficial fusion—it achieves top-tier accuracy across diverse clinical imaging tasks. Coupled with fast sampling via DPM-Solver, modular design, and strong community support, it’s a compelling choice for researchers and developers seeking a modern, high-performance segmentation framework that’s both scientifically rigorous and practically deployable. If your work involves extracting precise anatomical or pathological boundaries from medical scans, MedSegDiff deserves a serious look.